The AP2 team has isolated and preserved over 2,000 microbes from two sites in Sulawesi, Indonesia. Identification and relocation of microbes to the US is underway. Microbes are being screened by the AP2 team for enzyme activity and high lipid content.
Microbe Survey
The goals of the microbe survey are:
- Isolate microbes to be used by the AP3 team for therapeutic screening, including filamentous fungi and bacteria (including actinomycetes) from soil and plant surfaces.
- Isolate microbes from the guts of wood-feeding beetle larvae, and decaying wood and other plant matter, for cellulase activity screening.
- Isolate microbes from plant surfaces to screen for high lipid content, as a renewable source of oleochemicals for bulk and fine chemicals including fuels, adhesives, polymers, etc.
To date, we have isolated over 2,000 microbes from Sulawesi. Specimens have been collected at the Mekongga, Papalia, Rawa Aopa and Mangolo sites. We have found many interesting microbes, including close to 50 novel species of yeasts!
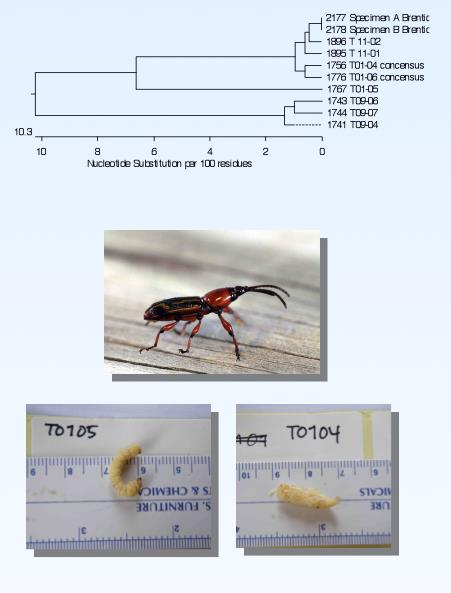
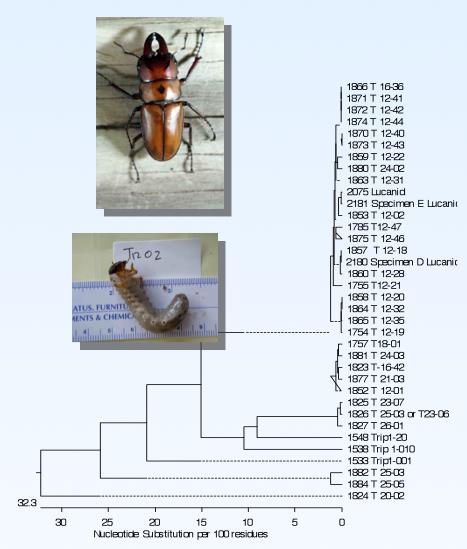
An understanding of the microbial ecology of the insect larva gut requires that we identify the larval species. This is technically difficult, as larvae lack the morphological features used to identify beetles: legs, wings, antennae. To get around this problem, we are performing DNA sequencing on tissue samples collected from the larvae we dissect, as well as adult insects collected at the same location. We have been able to match larval and adult DNA sequences to identify our larvae species.
Bioenergy Screening
Our bioenergy screening targets two technologies: improved cellulolytic enzymes, and high lipid (oleaginous) microbes.
Cellulase screening
Fungi (both filamentous fungi and yeasts) are being screened for ability to break down carboxymethyl cellulose (CMC) in agar, and to hydrolyze AZCL-conjugated substrates including AZCL-xylan and AZCL-arabinan. We have found many active fungi on both of these assays. We are now amplifying fragments of xylanase genes from a variety of fungi.

The search for oleaginous yeasts
A few oleaginous (high lipid) yeasts such as Yarrowia, Rhodotorula and Lipomyces have been studied for many years as a potential source of oleochemicals. Some of these species are reported to accumulate over 60% oil by dry weight! Most yeast species were not previously analyzed to determine how much oil they can accumulate. Our goals are to evaluate the oil content of a broad range of yeast species in order to find new oleaginous species, and then to determine which yeasts are best suited for conversion of different plant hydrolysates to oil.
An improved fluorescent assay for evaluating yeast cell lipid content was published in 2012. Upcoming publications include a survey of lipid content of 50 potentially oleaginous yeast species when grown under three different conditions, and then a report of their ability to utilize the sugars and tolerate the toxins found in lignocellulosic hydrolysates.